3第四代测序技术
纳米孔测序技术虽属单分子测序,但不同于Heli Scope遗传分析系统和Pac Bio RS单分子实时测序平台,它无需进行合成反应、荧光标记、洗脱和电荷耦合器件 (charge couple device,CCD) 照相机摄像,实现了从光学检测到电子传导检测和短读长到长读长测序的双重跨越,为真正的单分子测序技术,故归为第四代测序技术。英国OxfoldNanopore Technologies公 司 现 已 推 出 高 通 量 的Grid ION(2012年) 和U盘大小的Min ION(2013年) 测序仪[31],它们均处于试用阶段。瑞士Roche公司、美国Illumina和Life Technologies等公司也在投资纳米孔测序,如Roche投资了Genia Tech-nologies和Stratos Genomics公司[32].与其他NGS平台相比,纳米孔测序技术具长读长、高通量、低成本、短耗时和数据分析相对简单的优势,未来纳米孔测序技术投入市场后有望在几小时、几百美元的成本内完成全基因组测序。
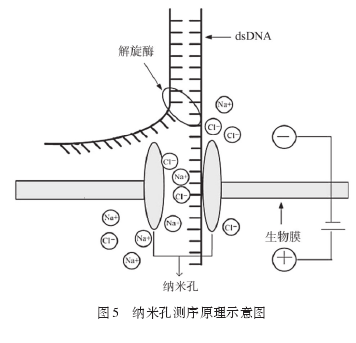
纳米孔测序原理见图5.由于DNA分子每个碱基的大小形状不同,DNA分子在电泳驱动下通过纳米微孔组成电路时可引起特征性电流变化,据此可确定DNA分子的碱基类型和排列顺序。纳米孔有生物纳米孔和固态纳米孔两种[33],生物纳米孔有α-溶血素纳米孔和Msp A蛋白纳米孔[34-35],固态纳米孔如石墨烯纳米孔[36].与生物纳米孔相比,固态纳米孔稳定性更好。Min ION成本较低,U盘大小,读长可达10 kb[37],但DNA分子通过速度快,难以区分碱基信息和本底噪音[38],错误率很高[39],因此还有待改进。在应用方面,纳米孔测序可用于单核苷酸、ss DNA、ds DNA和RNA测序,同时纳米孔技术还用于DNA、微小RNA、蛋白质、阴离子、阳离子和有机分子的定性与定量。
4结语
NGS平台的更新实现了全基因组的高通量、低成本和快速化检测,尤其是第二代测序技术的逐渐成熟,促进了NGS在临床领域的应用。越来越多的研究者选择NGS的基因panel检测 (一种芯片上含若干基因对应的探针来捕获目标DNA并进行基因测序的方法) 对肿瘤进行研究,或者通过外显子测序鉴定潜在的药物靶点。在诊断儿童遗传性疾病时,也逐渐抛开了传统的诊断方法,转而使用全外显子组测序。NGS检测既可以帮助医生确定治疗方案,又可以为患儿父母评估他们未来子女患同样疾病的风险,必将推动分子诊断革新和精准医疗的发展。总体而言,Illumina测序平台应用最为普遍,测序成本也显着降低,首次达到1 000美元完成人类基因组测序的目标;Roche 454因其较长的测序读长,主要用于从头测序和宏基因组学研究;ABI SOLi D比Illumina应用少;介于第二代和第三代间的Ion Torrent PGM,因其简单、快速、低成本和小规模优势而得到广泛应用。在应用于临床时,第二代测序的结果仍需要测序金标准--第一代测序技术的验证。第三代和第四代测序技术目前处于发展阶段,虽读长比Sanger测序法长十几倍,但Pac Bio RS单分子实时测序系统和Min ION测序仪错误率高,还有待改进。另外,NGS数据的生物信息学分析也面临着巨大挑战。总之,随着对基因组信息更深层次的理解、测序技术的进步和生物信息学分析软件的革新,测序技术将在生物学和生物医学研究中发挥更大的作用。
参考文献:
[1] Buermans H P, den Dunnen J T. Next generation sequenc-ing technology: advances and applications[J]. BiochimBiophys Acta, 2014, 1842(10): 1932-1941.
[2] Baudhuin L M. A new era of genetic testing and its im-pact on research and clinical care[J]. Clin Chem, 2012,58(6): 1070-1071.
[3] Chan L L, Jiang P. Bioinformatics analysis of circulatingcell- free DNA sequencing data[J]. Clin Biochem, 2015,48(15): 962-975.
[4] Meldrum C, Doyle M A, Tothill R W. Next- generation se-quencing for cancer diagnostics: a practical perspective[J].Clin Biochem Rev, 2011, 32(11): 177-195.
[5] Mestan K K, Ilkhanoff L, Mouli S, et al. Genomic se-quencing in clinical trials[J]. J Transl Med, 2011, 9: 222.
[6] Behjati S, Tarpey P S. What is next generation sequenc-ing[J]. Arch Dis Child Educ Pract Ed, 2013, 98(6): 236-238.
[7] Van Dijk E L, Auger H, Jaszczyszyn Y, et al. Ten years ofnext-generation sequencing technology[J]. Trends Genet,2014, 30(9): 418-426.
[8] Roche. Products[EB/OL]. [2016- 4- 7].
http://454.com/in-dex. asp.
[9] Xuan J, Yu Y, Qing T, et al. Next- generation sequencingin the clinic: promises and challenges[J]. Cancer Lett,2013, 340(2): 284-295.
[10] Morey M, Fernandez-Marmiesse A, Castineiras D, et al.A glimpse into past, present, and future DNA sequenc-ing[J]. Mol Genet Metab, 2013, 110(1-2): 3-24.
[11]华大基因。华大技术平台[EB/OL]. [2016- 4- 7].
http://www.genomics.cn/navigation/show_navigation?nid=220.
[12] Illimina. Systems[EB/OL]. [2016- 4- 7].
http://www.illu-mina.com/.
[13] Kampmann M L, Fordyce S L, Avila-Arcos M C, et al.A simple method for the parallel deep sequencing of fullinfluenza A genomes [J]. J Virol Methods, 2011,78(8):243-248.
[14] Golan D, Medvedev P. Using state machines to modelthe ion torrent sequencing process and to improve readerror rates[J]. Bioinformatics, 2013, 29(13): 344-351.
[15] Jessri M, Farah C S. Next generation sequencing and itsapplication in deciphering head and neck cancer[J]. OralOncology, 2014, 50 (4): 247-253.
[16] Salipante S J, Kawashima T, Rosenthal C, et al. Perfor-mance comparison of illumina and ion torrent next- gen-eration sequencing platforms for 16S r RNA-based bacte-rial community profilin[J]. Appl Environ Microbiol,2014, 80(24): 7583-7591.
[17] Daniel R, Santos C, Phillips C, et al. A SNa Pshot of nextgeneration sequencing for forensic SNP analysis[J].Forensic Sci Int Genet, 2015, 14(6): 50-60.
[18] Fordyce S L, Mogensen H S, Borsting C, et al. Second-generation sequencing of forensic STRs using the ion tor-rent HID STR 10-plex and the Ion PGM[J]. Forensic SciInt Genet, 2015, 14: 132-140.
[19] Paparini A, Gofton A, Yang R, et al. Comparison ofSanger and next generation sequencing performance forgenotyping cryptosporidium isolates at the 18S r RNAand actin loci[J]. Exp Parasitol, 2015, 151: 21-27.
[20] Parson W, Strobl C, Huber G, et al. Evaluation of nextgeneration mt Genome sequencing using the ion torrentpersonal genome machine (PGM) [J]. Forensic Sci IntGenet, 2013, 7(5): 543-549.
[21] Chen S, Li S, Xie W, et al. Performance comparison be-tween rapid sequencing platforms for ultra-low coveragesequencing strategy[J]. PLo S One, 2014, 9(3): e92192.
[22] Schadt E E, Turner S, Kasarskis A. A window into third-generation sequencing[J]. Hum Mol Genet, 2010, 19(R2):R227-240.
[23] Korlach J, Turner S W. Going beyond five bases in DNAsequencing[J]. Curr Opin Struct Biol, 2012, 22(3): 251-261.
[24] Thompson J F, Steinmann K E. Single molecule sequenc-ing with a Heli Scope Genetic Analysis System[M]. CurrProtoc Mol Biology. John Wiley & Sons, 2010: 7.10.1-7.10.14.
[25] Orlando L, Ginolhac A, Raghavan M, et al. True single-molecule DNA sequencing of a pleistocene horse bone[J].Genome Res, 2011, 21(10): 1705-1719.
[26] Yu N Y, Hallstrom B M, Fagerberg L, et al. Complement-ing tissue characterization by integrating transcriptomeprofiling from the human protein atlas and from the FAN-TOM5 consortium[J]. Nucleic Acids Res, 2015, 43(14):6787-6798.
[27] Itoh M, Kojima M, Nagao-Sato S, et al. Automated work-flow for preparation of c DNA for cap analysis of gene ex-pression on a single molecule sequencer[J]. PLo S One,2012, 7(1): e30809.
[28] Mc Ginn S, Gut I G. DNA sequencing-spanning the gen-erations[J]. N Biotechnol, 2013, 30(4): 366-372.
[29] Faino L, Thomma B P. Get your high- quality low- costgenome sequence[J]. Trends Plant Sci, 2014, 19(5): 288-291.
[30] Rhoads A, Au K F. Pac Bio sequencing and its appli-cations[J]. Genomics Proteomics Bioinformatics, 2015,13(5): 278-89.
[31] Feng Y, Zhang Y, Ying C, et al. Nanopore-based fourth-generation DNA sequencing technology[J]. GenomicsProteomics Bioinformatics, 2015, 13(1): 4-16.
[32]仪器信息网。从二代到四代:基因测序颠覆你对世界的“想象力”[EB/OL]. (2015-3-3)[2016-4-7].
http://www.instrument.com.cn/news/20150303/154348.shtml.
[33] Mattei T A. Biomimetic nanopores: a new age of DNA se-quencing[J]. World Neurosurg, 2014, 82(3-4): 252-254.
[34] Manrao E A, Derrington I M, Laszlo A H, et al. ReadingDNA at single- nucleotide resolution with a mutantMsp A nanopore and phi29 DNA polymerase[J]. NatBiotechnol, 2012, 30(4): 349-353.
[35] Haque F, Li J, Wu H C, et al. Solid- state and biologicalnanopore for real-time sensing of single chemical and se-quencing of DNA[J]. Nano Today, 2013, 8(1): 56-74.
[36] Stoloff D H, Wanunu M. Recent trends in nanopores forbiotechnology[J]. Curr Opin Biotechnol, 2013, 24(4):699-704.
[37] Laver T, Harrison J, O'Neill P A, et al. Assessing theperformance of the Oxford Nanopore TechnologiesMin ION[J]. Biomol Detect Quantif, 2015, 3: 1-8.
[38] Luan B, Stolovitzky G, Martyna G. Slowing and con-trolling the translocation of DNA in a solid- state nano-pore[J]. Nanoscale, 2012, 4(4): 1068-1077.
[39] Mikheyev A S, Tin M M. A first look at the OxfordNanopore Min ION sequencer[J]. Mol Ecol Resour,2014, 14(6): 1097-1102.